Why Every Health Data Scientist Should Know About OMOP CDM
Standardising Healthcare Data
A Key Problem with Healthcare Data
A large issue I struggle with at work is standardising healthcare data.
I gather data from hospitals around the world in an attempt to produce a centralised and unified database for participating hospitals and researchers to benefit from.
Each hospital seems to have it’s own way of recording healthcare data. There are many standards out there which hospitals follow but there is no universal agreement on how healthcare data should be recorded. As a result, I find myself mapping different data collection standards to fit a standard produced by my organisation.
There are many domains in medicine, each of which have specific variables that need to be collected. For example cardiology data collection requires the collection of different variables than oncology or endocrinology. These domain-specific requirements adds a layer of complexity to standardisation, not only do we need to align general health data such as age, sex and BMI, but we must also ensure that the needs of each speciality is sufficiently met.
Producing a data model that has the ability to accurately capture information from every healthcare domain is an immense challenge. However, if this challenge is met and the resulting model is widely adopted this would lead to a scalable and interoperable solution that enables data across healthcare systems to be unified without much effort. The Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM) was developed to address this specific challenge.
Supplementary Video
Building a Common Language for Healthcare Data
OMOP CDM is a relational database model that enables the conversion of healthcare data into a consistent format. It’s about making a common language so diverse datasets can be combined and analysed more easily, unifying different ways of recording medical observations. To achieve this, Observational Health Data Sciences and Informatics (OHDSI) developed standardised vocabularies which allow the organisation and standardisation of medical terms, some gathered from globally recognised coding systems such as ICD-10, SNOMED-CT, and RxNorm, to be used across the clinical domains of OMOP CDM.
For example, pancreatic cancer has the code 648 which is under the Domain ID “Condition” and Aspirin has the code 387458008 which is under the Domain ID “Drug”. Whether it is a condition, device, drug or procedure — each medical observation is captured with a specific set of codes which are then utilised in OMOP CDM. This structure is useful because if a new drug or disease is discovered a new code can be assigned to this and recorded in OMOP CDM.
A full list of the vocabularies can be viewed here:
https://athena.ohdsi.org/search-terms/start
OMOP CDM consists of 37 tables and 394 fields with the following structure:
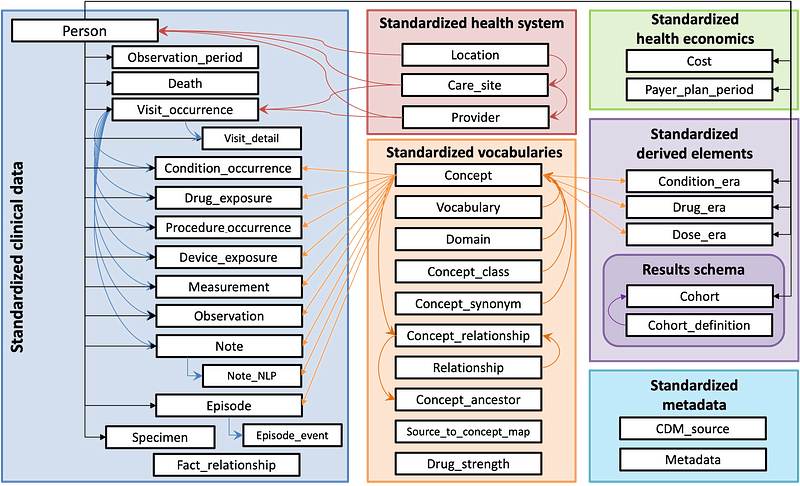
Data can be standardised and grouped into 7 sections:
Clinical Data: Captures patient-specific clinical information such as diagnoses, medication and procedures.
Health System: Represents the healthcare infrastructure.
Health Economics: Stores financial and insurance-related data.
Standardised Vocabularies: Mentioned earlier in the article, holds standardised medical terminology and relationships to ensure consistent data across various sources.
Derived Elements: Includes derived data such as the duration of condition or drug exposures.
Results Schema: Contains information for cohort analysis which supports the definition and grouping of patient populations for research studies.
Metadata: Stores metadata about the data source and the OMOP CDM instance to help document and track the data’s origin and structure.
Now that we have a brief overview of what OMOP CDM is and it’s purpose, let us look into some of the reasons as to why this data model matters.
Why OMOP CDM Matters
1. Making Healthcare Data Interoperable and Scalable
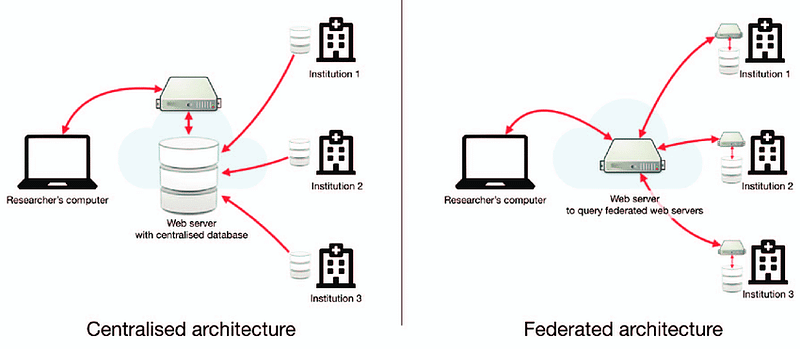
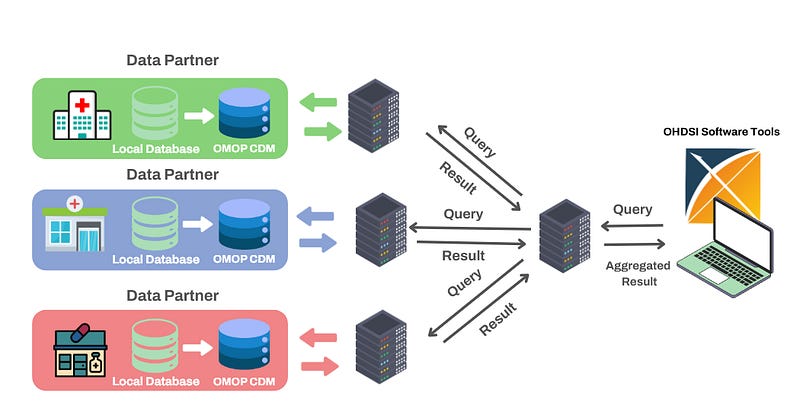
The above image gives examples of two common data architectures that can be adopted to enable researchers to access and analyse data gathered from different institutions.
Federated architecture is particularly well suited to the healthcare industry. As demonstrated in the above image, each database remains within it’s originating institution which ensures that sensitive patient information is never shared or transferred beyond the hospital’s control. Each hospital retains full ownership of their data which reduces concerns about potential breaches or unauthorised access and aligns well with data protection standards such as GDPR.
In addition, this architecture enables something called federated learning, where analyses can be performed from each institution’ s database without the need of a centralised database. This is done by aggregating results from each institution via a web server.
This federated architecture has been initiated by EHDEN (https://www.ehden.eu/datapartners/) which currently have 187 data partners from 29 different countries that are mapping their data to the OMOP common data model:
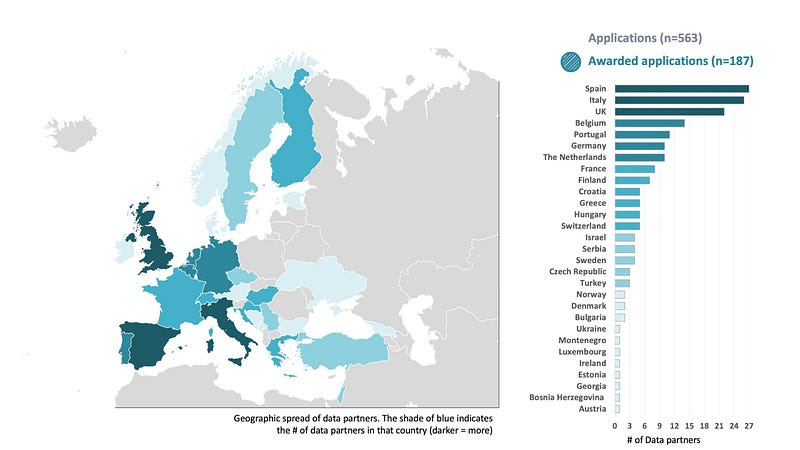
By establishing this architecture EHDEN are providing an incredible resource to researchers for deriving new insights from standardised medical data and hospitals that wish to monitor and enhance their current practises.
Standardising Healthcare Analysis
A list of open-source software tools have been built by the Observational Health Data Sciences and Informatics (OHDSI) community to enable various standardised analyses from participating organisations or data partners that have adopted OMOP CDM. Examples of these tools include:
ATLAS: A powerful web-based platform that provides a user-friendly interface for defining patient cohorts, summarise characteristics about these patient populations, estimate incidence rates for populations of interest and apply machine learning algorithms to conduct patient level prediction analyses whereby you can predict an outcome within any given target exposures. ATLAS enables clinicians and researchers to generate valuable insights from OMOP CDM compliant datasets without requiring extensive programming skills.
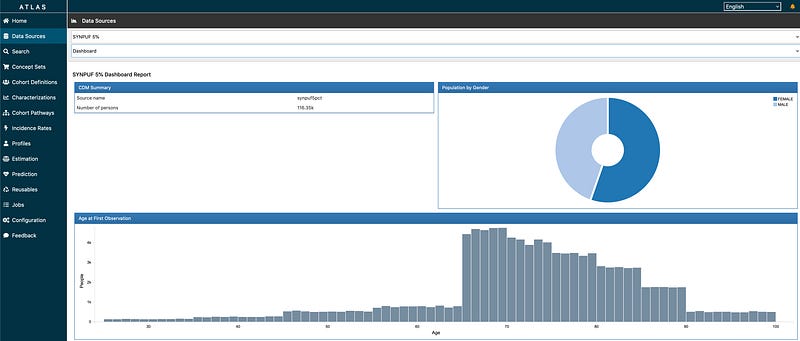
Read more about ATLAS from The Book of OHDSI Chapter 8.3 here: https://ohdsi.github.io/TheBookOfOhdsi/OhdsiAnalyticsTools.html#atlas
HAEDES: The Health Analytics Data-to-Evidence Suite is a collection of R packages that were developed to facilitate observational research using OMOP CDM. HADES provides advances standardised analytics for population characterisation, population level-effect estimation and patient-level prediction. By offering a robust and reproducible set of tools, HADES aids in streaming the analytics workflow and ensures consistency in health data research. Users can also use these standardised packages to help with completely custom analyses.

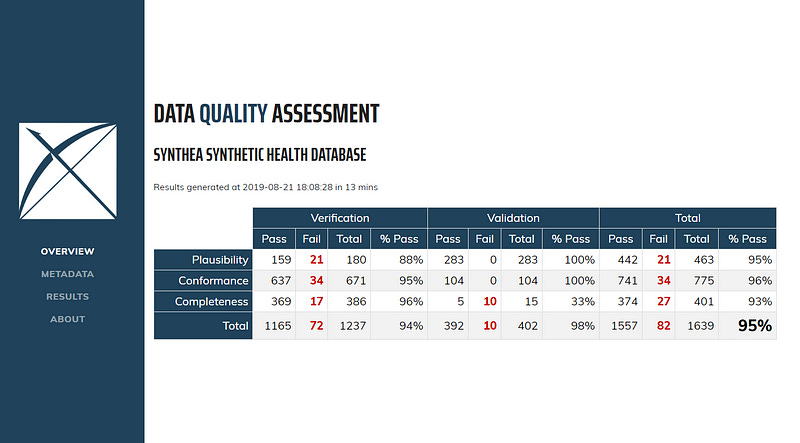
Data Quality Dashboard (DQD): Healthcare research requires high-quality data to prevent erroneous results. DQD was developed to help meet this requirement by performing over 1,500 automated quality checks on OMOP CDM-compliant databases according to the Kahn framework. This framework addresses three key areas:
Conformance: Do data values adhere to specified standards and formats?
Completeness: Whether a particular variable is present as well as whether variables contain all recorded values.
Plausibility: Are data values believable?
Read more about DQD from The Book of OHDSI Chapter 15.5 here:
https://ohdsi.github.io/TheBookOfOhdsi/DataQuality.html#ref-kahn_harmonized_2016
Accelerating AI and Machine Learning
OMOP CDM is really a treasure for health data scientists and machine learning engineers that are looking to construct new models for predictive analytics or implement unsupervised techniques for knowledge discovery. By providing a standardised and interoperable data structure, OMOP CDM makes it much easier to gather and integrate data from diverse sources, which is crucial for building robust machine learning models.
Mentioned in the above section, the OHDI community have developed a set of software tools to support AI and machine learning endeavours. ATLAS can be used to define patient cohorts which can be used as input data for training predictive models. The user-friendly platform can help health data scientists to focus more on model development rather than data wrangling.
A wide array of off the shelf machine learning models can be used from HADES to implement and evaluate predictive models for user-specified outcomes. This toolset promotes transparency and reproducibility which are essential components of modern healthcare research.
Getting Started with OMOP CDM
If you are an institution that collects medical data and would like to have your data available in OMOP CDM format to help drive global collaboration and research there are a few important tools to help you with this process.
ATHENA is a repository that manages OMOP CDM standardised medical vocabularies for concepts such as procedures, diagnoses and drugs. With ATHENA you can search through OMOP terminologies to ensure your data uses the correct codes.
Athena
Edit descriptionathena.ohdsi.org
To help map the codes in your data to OMOP’s standard concepts you can make use of USAGI. USAGI is a tool that uses natural language processing (NLP) to suggest matches between your local codes and OMOP standards. While this platform provides automated suggestions, it is important to manually review these matches to ensure accuracy and make any necessary adjustments.
To gain an understanding of what needs to be transformed from your local data to OMOP CDM use WhiteRabbit. WhiteRabbit performs a scan of the source data and provides detailed information on the tables, fields and values that appear in a field. It differs from standard data profiling tools by attempting not to display patient identifiable information (PII) in the scan report. The scan report produced by WhiteRabbit can then be used in conjunction with Rabbit-in-a-Hat to display a user interface that allows you to connect source data to tables and columns in OMOP CDM.
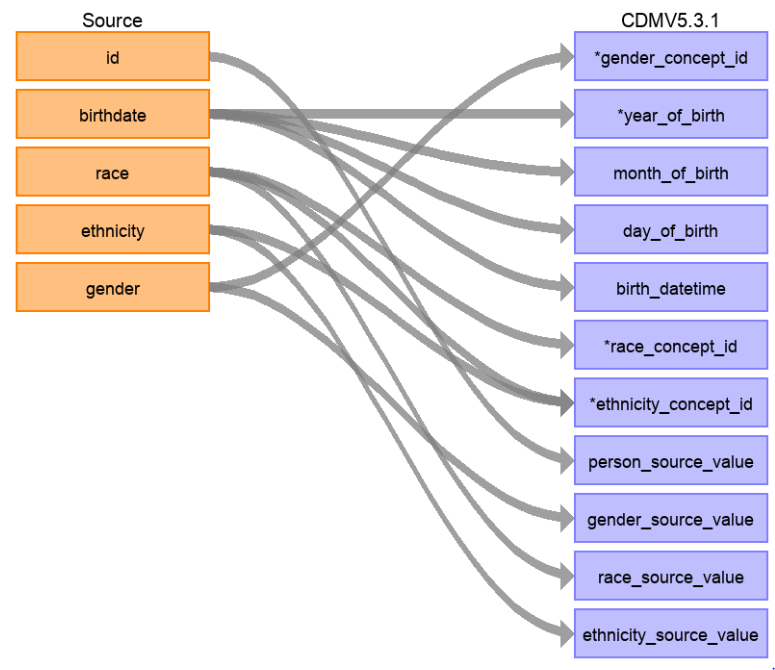
Each of these tools can be used to help with the transformation process from your local source data to OMOP CDM. It is important to not start this process until your design is complete.
The Future of OMOP CDM
I am hopeful to see the continued global adoption and expansion of OMOP CDM, which has already gained significant traction across multiple countries and healthcare systems. The push for a unified global healthcare data model can enable larger scale multinational studies and provide stronger evidence for clinical practises and policies.
As OMOP CDM continues to develop it has the potential to expand beyond healthcare and into related fields such as public health, social care and environmental health. By integrating data from these sectors we provide a more holistic view on healthcare. This expansion could facilitate research that looks at the interplay between clinical care, social factors and environmental health. I do see the integration of this data to be an incredible challenge, however, as much of this information would have to be gathered and linked to data from medical institutions.
I see strong progress in the toolset developed by OHDSI for standardised analytics and mapping source data to OMOP CDM. These mapping tools currently require a fair amount of manual intervention. In the future, we can expect further automation and streamlining of these processes which would allow smaller healthcare organisations and researchers with limited resources to transform their data to OMOP CDM and be a part of the global network.
Let’s Connect!
References
[1] OHDSI. (n.d.). Data Standardization — OHDSI. [online] Available at: https://www.ohdsi.org/data-standardization/.
[2]ehden.eu. (2024). Data Partners — ehden.eu. [online] Available at: https://www.ehden.eu/datapartners/ [Accessed 27 Oct. 2024].
[3] ResearchGate. (2024). Figure 2. Comparison of centralized versus federated architectures. In… [online] Available at: https://www.researchgate.net/figure/Comparison-of-centralized-versus-federated-architectures-In-the-centralized_fig2_334205057 [Accessed 28 Oct. 2024].
[4] Kahn, Michael G., Tiffany J. Callahan, Juliana Barnard, Alan E. Bauck, Jeff Brown, Bruce N. Davidson, Hossein Estiri, et al. 2016. “A Harmonized Data Quality Assessment Terminology and Framework for the Secondary Use of Electronic Health Record Data.” EGEMS (Washington, DC) 4 (1): 1244. https://doi.org/10.13063/2327-9214.1244.
[5] Anon, (n.d.). Software Tools — OHDSI. [online] Available at: https://www.ohdsi.org/software-tools/.
[6] Ohdsi.org. (2022). Building Your CDM — OHDSI. [online] Available at: https://www.ohdsi.org/data-standardization/building-your-cdm/ [Accessed 29 Oct. 2024].